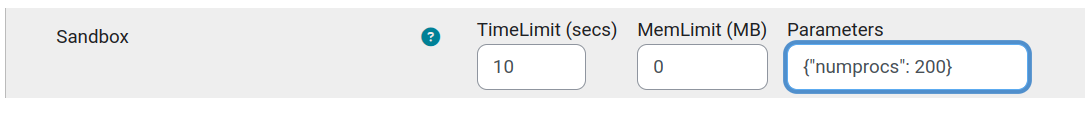
There is something very odd about your Jobe server configuration.
With some help from my AI mate Claude, here's a Python script for you to run via CodeRunner [**EDIT** Code changed to fix bug in reporting of resource limits and to avoid the use of the psutil module]. Please report its output:
#!/usr/bin/env python3
import os
import platform
import subprocess
import sys
def get_memory_info():
"""Get memory information from /proc/meminfo"""
try:
with open('/proc/meminfo') as f:
meminfo = {}
for line in f:
key, value = line.split(':')
# Convert kb to bytes and remove ' kB' suffix
value = int(value.strip().split()[0]) * 1024
meminfo[key] = value
total_gb = round(meminfo['MemTotal'] / (1024**3), 2)
available_gb = round((meminfo.get('MemAvailable', meminfo['MemFree'])) / (1024**3), 2)
used_percent = round(((meminfo['MemTotal'] - meminfo.get('MemAvailable', meminfo['MemFree'])) /
meminfo['MemTotal']) * 100, 2)
return total_gb, available_gb, used_percent
except:
return None, None, None
def get_cpu_info():
"""Get CPU information from /proc/cpuinfo"""
try:
with open('/proc/cpuinfo') as f:
cpuinfo = f.read()
physical_ids = set()
cpu_cores = set()
for line in cpuinfo.split('\n'):
if line.startswith('physical id'):
physical_ids.add(line.split(':')[1].strip())
elif line.startswith('processor'):
cpu_cores.add(line.split(':')[1].strip())
return len(physical_ids) or 1, len(cpu_cores)
except:
return None, None
def get_disk_usage(path='/'):
"""Get disk usage information"""
try:
st = os.statvfs(path)
total = st.f_blocks * st.f_frsize
free = st.f_bfree * st.f_frsize
return round(total / (1024**3), 2), round(free / (1024**3), 2)
except:
return None, None
def get_system_info():
info = {}
# Basic system info
info['python_version'] = sys.version
info['platform'] = platform.platform()
info['architecture'] = platform.machine()
info['processor'] = platform.processor()
# Memory information
total_mem, available_mem, mem_percent = get_memory_info()
if total_mem is not None:
info['total_memory_gb'] = total_mem
info['available_memory_gb'] = available_mem
info['memory_percent_used'] = mem_percent
# CPU information
physical_cpus, logical_cpus = get_cpu_info()
if physical_cpus is not None:
info['cpu_count_physical'] = physical_cpus
info['cpu_count_logical'] = logical_cpus
# Linux-specific information
if platform.system() == 'Linux':
try:
# Get Linux distribution details
with open('/etc/os-release') as f:
for line in f:
if line.startswith('PRETTY_NAME='):
info['linux_distribution'] = line.split('=')[1].strip().strip('"')
break
# Check SELinux status
try:
selinux_status = subprocess.check_output(['getenforce'], text=True).strip()
info['selinux_status'] = selinux_status
except (subprocess.CalledProcessError, FileNotFoundError):
info['selinux_status'] = 'Not installed or not accessible'
# Get available storage
root_total, root_free = get_disk_usage('/')
if root_total is not None:
info['root_total_gb'] = root_total
info['root_free_gb'] = root_free
# Check for common limiting factors
# Max processes
try:
with open('/proc/sys/kernel/pid_max') as f:
info['max_processes'] = f.read().strip()
except:
info['max_processes'] = 'Unable to determine'
# Resource limits
try:
ulimit_output = subprocess.check_output(['bash', '-c', 'ulimit -a'],
shell=False, text=True)
info['resource_limits'] = ulimit_output.strip()
except:
info['resource_limits'] = 'Unable to determine'
# Check for containerization
try:
with open('/proc/1/cgroup') as f:
cgroup_content = f.read()
info['containerized'] = ('docker' in cgroup_content.lower() or
'lxc' in cgroup_content.lower())
except:
info['containerized'] = 'Unable to determine'
except Exception as e:
info['linux_specific_error'] = str(e)
return info
def format_output(info):
output = "=== System Diagnostics ===\n\n"
# Format the output nicely
for key, value in info.items():
if key == 'resource_limits':
output += f"\n=== Resource Limits ===\n{value}\n"
else:
output += f"{key.replace('_', ' ').title()}: {value}\n"
return output
if __name__ == "__main__":
try:
info = get_system_info()
print(format_output(info))
except Exception as e:
print(f"Error gathering system information: {e}")